Last modified: March 14, 2024
This article is written in: 🇺🇸
Anomaly Detection in Machine Learning
Anomaly detection involves identifying data points that significantly differ from the majority of the data, often signaling unusual or suspicious activities. This technique is widely used across various domains, such as fraud detection, manufacturing, and system monitoring.
Concept of Anomaly Detection
- Baseline Establishment: The first step in anomaly detection is to establish a normal pattern or baseline from the dataset.
- Probability Threshold (Epsilon): Data points are flagged as anomalies if their probability, according to the established model, falls below a threshold $\epsilon$.
- If $p(x_{\text{test}}) < \epsilon$, the data point is considered an anomaly.
- If $p(x_{\text{test}}) \geq \epsilon$, the data point is considered normal.
- Threshold Selection: The value of $\epsilon$ is critical and is chosen based on the desired confidence level and the specific context of the application.
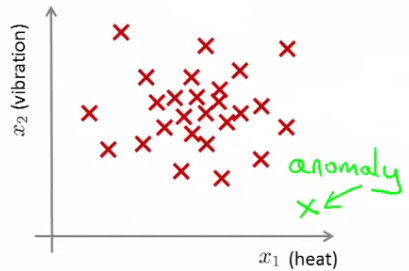
Applications of Anomaly Detection
I. Fraud Detection:
- User behavior metrics (online time, login location, spending patterns) are analyzed.
- A model of typical user behavior is created, and deviations are flagged as potential fraud.
II. Manufacturing: In scenarios like aircraft engine production, anomalies can indicate defects or potential failures.
III. Data Center Monitoring: Monitoring metrics (memory usage, disk accesses, CPU load) to identify machines that are likely to fail.
Utilizing the Gaussian Distribution
- Mean ($\mu$) and Variance ($\sigma^2$): The Gaussian distribution is characterized by these parameters.
- Probability Calculation:
$$ p(x; \mu; \sigma^2) = \frac{1}{\sqrt{2\pi\sigma^2}} \exp\left(-\frac{(x-\mu)^2}{2\sigma^2}\right) $$
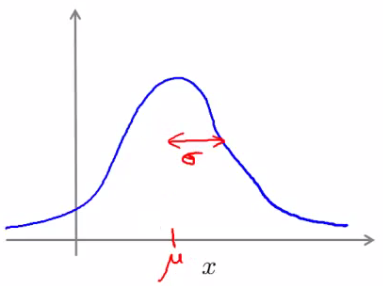
- Data Fitting: Estimating the Gaussian distribution from the dataset to represent the normal behavior.
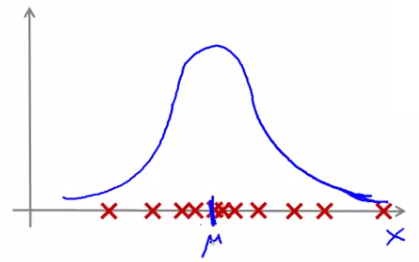
Evaluating Anomaly Detection Systems
- Data Split: Divide the data into training, cross-validation, and test sets.
- Performance Metrics: Precision and recall are often used to evaluate the effectiveness of the anomaly detection system.
- Fine-Tuning: Adjust the model and $\epsilon$ based on the performance metrics to improve accuracy.
Algorithm Steps
-
Feature Selection: Choose features $x_i$ that might be indicators of anomalous behavior.
-
Parameter Fitting: Calculate the mean ($\mu_j$) and variance ($\sigma_j^2$) for each feature
$$ \mu_j = \frac{1}{m} \sum_{i=1}^m x_j^{(i)} $$
$$ \sigma_j^2 = \frac{1}{m} \sum_{i=1}^m (x_j^{(i)} - \mu_j)^2 $$
- Probability Computation for New Example: For a new example $x$, compute the probability $p(x)$:
$$ p(x) = \prod_{j=1}^n \frac{1}{\sqrt{2\pi\sigma_j^2}} \exp\left(-\frac{(x_j - \mu_j)^2}{2\sigma_j^2}\right) $$
Below is the Python implementation of the Gaussian distribution, data fitting, and evaluation of anomaly detection systems based on the algorithm steps.
import numpy as np
# Function to calculate Gaussian Probability
def gaussian_probability(x, mean, variance):
coefficient = 1 / np.sqrt(2 * np.pi * variance)
exponent = np.exp(-((x - mean) ** 2) / (2 * variance))
return coefficient * exponent
# Function to fit Gaussian parameters (mean and variance) for each feature
def fit_gaussian_parameters(X):
mean = np.mean(X, axis=0)
variance = np.var(X, axis=0)
return mean, variance
# Function to calculate the probability of a new example using the Gaussian distribution
def compute_probability(x, mean, variance):
probabilities = gaussian_probability(x, mean, variance)
return np.prod(probabilities)
# Example dataset
X_train = np.array([[1.1, 2.2], [1.3, 2.1], [1.2, 2.3], [1.1, 2.4]])
X_cross_val = np.array([[1.0, 2.0], [1.4, 2.5]])
# Fitting the Gaussian parameters
mean, variance = fit_gaussian_parameters(X_train)
# Calculate the probability of a new example
x_new = np.array([1.2, 2.2])
probability = compute_probability(x_new, mean, variance)
print("Mean:", mean)
print("Variance:", variance)
print("Probability of new example:", probability)
# Performance Metrics (Precision and Recall)
# Note: This requires true labels and predicted labels
from sklearn.metrics import precision_score, recall_score
# Example true labels and predicted labels
y_true = np.array([0, 0, 1, 1])
y_pred = np.array([0, 0, 1, 0])
precision = precision_score(y_true, y_pred)
recall = recall_score(y_true, y_pred)
print("Precision:", precision)
print("Recall:", recall)Here are the results:
I. Gaussian Parameters:
- Mean: Feature 1: 1.175, Feature 2: 2.25
- Variance: Feature 1: 0.006875, Feature 2: 0.0125
II. Probability of New Example:
- For the new example $[1.2, 2.2]$, the calculated probability using the Gaussian distribution is approximately 14.844.
III. Performance Metrics:
- Precision: 1.0 (All instances predicted as anomalies are actually anomalies, with no false positives)
- Recall: 0.5 (Only 50% of the actual anomalies were correctly identified, indicating some false negatives)
Developing and Evaluating an Anomaly Detection System
I. Labeled Data: Have a dataset where $y=0$ indicates normal (non-anomalous) examples, and $y=1$ represents anomalous examples.
II. Data Division: Separate the dataset into a training set (normal examples), a cross-validation (CV) set, and a test set, with both the CV and test sets including some anomalous examples.
III. Example Case:
- Imagine a dataset with 10,000 normal (good) engines and 50 flawed (anomalous) engines.
- Training set: 6,000 good engines.
- CV set: 2,000 good engines, 10 anomalous.
- Test set: 2,000 good engines, 10 anomalous.
IV. Evaluation Metrics:
- True positives (TP), false positives (FP), false negatives (FN), and true negatives (TN).
- Precision (the proportion of true positives among all positives identified by the model).
- Recall (the proportion of true positives identified out of all actual positives).
- F1-score (a harmonic mean of precision and recall, providing a balance between them).
Below is the complete Python code for the implementation:
import numpy as np
from sklearn.metrics import precision_score, recall_score, f1_score
# Simulate dataset
np.random.seed(0)
# Normal examples
normal_examples = np.random.normal(0, 1, (10000, 2))
# Anomalous examples
anomalous_examples = np.random.normal(5, 1, (50, 2))
# Labels
y_normal = np.zeros(10000)
y_anomalous = np.ones(50)
# Combine the data
X = np.vstack((normal_examples, anomalous_examples))
y = np.concatenate((y_normal, y_anomalous))
# Shuffle the dataset
indices = np.arange(X.shape[0])
np.random.shuffle(indices)
X = X[indices]
y = y[indices]
# Data division
X_train = X[:6000]
y_train = y[:6000]
X_cv = X[6000:8000]
y_cv = y[6000:8000]
X_test = X[8000:]
y_test = y[8000:]
# Fit Gaussian parameters on the training set
mean, variance = fit_gaussian_parameters(X_train)
# Probability threshold
epsilon = 0.01 # This threshold can be tuned
# Compute probabilities for CV and Test sets
p_cv = np.array([compute_probability(x, mean, variance) for x in X_cv])
p_test = np.array([compute_probability(x, mean, variance) for x in X_test])
# Predict anomalies
y_pred_cv = (p_cv < epsilon).astype(int)
y_pred_test = (p_test < epsilon).astype(int)
# Calculate evaluation metrics for the CV set
tp_cv = np.sum((y_cv == 1) & (y_pred_cv == 1))
fp_cv = np.sum((y_cv == 0) & (y_pred_cv == 1))
fn_cv = np.sum((y_cv == 1) & (y_pred_cv == 0))
tn_cv = np.sum((y_cv == 0) & (y_pred_cv == 0))
precision_cv = precision_score(y_cv, y_pred_cv)
recall_cv = recall_score(y_cv, y_pred_cv)
f1_cv = f1_score(y_cv, y_pred_cv)
# Calculate evaluation metrics for the Test set
tp_test = np.sum((y_test == 1) & (y_pred_test == 1))
fp_test = np.sum((y_test == 0) & (y_pred_test == 1))
fn_test = np.sum((y_test == 1) & (y_pred_test == 0))
tn_test = np.sum((y_test == 0) & (y_pred_test == 0))
precision_test = precision_score(y_test, y_pred_test)
recall_test = recall_score(y_test, y_pred_test)
f1_test = f1_score(y_test, y_pred_test)
# Display results
results = {
"CV Set": {
"TP": tp_cv,
"FP": fp_cv,
"FN": fn_cv,
"TN": tn_cv,
"Precision": precision_cv,
"Recall": recall_cv,
"F1 Score": f1_cv
},
"Test Set": {
"TP": tp_test,
"FP": fp_test,
"FN": fn_test,
"TN": tn_test,
"Precision": precision_test,
"Recall": recall_test,
"F1 Score": f1_test
}
}
import ace_tools as tools; tools.display_dataframe_to_user(name="Anomaly Detection Results", dataframe=results)Below are the results for the cross-validation (CV) set and the test set.
I. Cross-Validation (CV) Set:
- True Positives (TP): 6
- False Positives (FP): 18
- False Negatives (FN): 4
- True Negatives (TN): 1982
- Precision: 0.25
- Recall: 0.6
- F1 Score: 0.35294117647058826
II. Test Set:
- True Positives (TP): 3
- False Positives (FP): 20
- False Negatives (FN): 7
- True Negatives (TN): 1980
- Precision: 0.13043478260869565
- Recall: 0.3
- F1 Score: 0.1818181818181818
III. Discussion:
- The precision is quite low in both the CV and test sets, indicating a high number of false positives. This means the model is predicting too many normal examples as anomalies.
- The recall is moderate in the CV set (0.6) but lower in the test set (0.3). This means the model is missing some anomalies, especially in the test set.
- The F1 score, which balances precision and recall, is also low, indicating the overall performance of the model needs improvement.
IV. Next Steps:
- Adjusting the threshold for anomaly detection could help balance precision and recall. A lower threshold might reduce false positives, while a higher threshold could reduce false negatives.
- Additional or better features could improve the model's ability to distinguish between normal and anomalous examples.
- Consider more sophisticated anomaly detection algorithms such as Isolation Forest, One-Class SVM, or neural networks designed for anomaly detection.
- Increasing the number of anomalous examples in the training set could help the model learn to identify anomalies more accurately.
Selecting a Good Evaluation Metric
- F1-Score: This is particularly useful in the context of anomaly detection where the class distribution is highly imbalanced (many more normal than anomalous examples).
- Precision and Recall: These metrics provide a more nuanced understanding of the system’s performance, especially when the cost of false positives and false negatives varies.
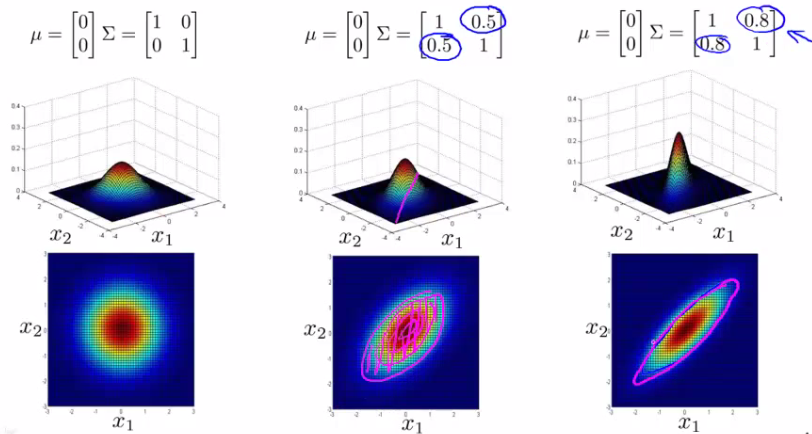
Concept of Multivariate Gaussian Distribution
- Scenario: Imagine fitting a Gaussian distribution to two features, such as CPU load and memory usage.
- Example: In a test set, consider an example with a high memory use (x1 = 1.5) but low CPU load (x2 = 0.4). Individually, these values might fall within normal ranges, but their combination could be anomalous.
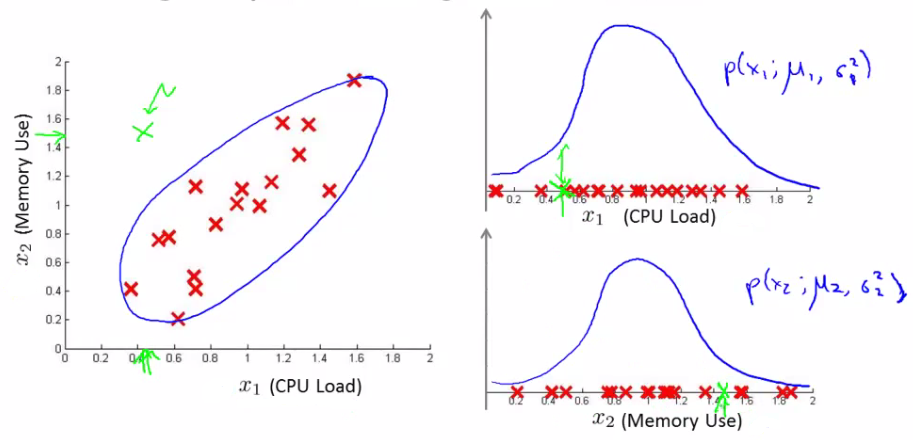
Parameters of the Multivariate Gaussian Model
- Mean Vector ($\mu$): An n-dimensional vector representing the mean of each feature.
- Covariance Matrix ($\Sigma$): An $[n \times n]$ matrix, capturing how each pair of features varies together.
- Probability Density Function:
$$ p(x; \mu; \Sigma) = \frac{1}{(2\pi)^{n/2}|\Sigma|^{1/2}} \exp\left(-\frac{1}{2}(x - \mu)^T \Sigma^{-1}(x - \mu)\right) $$
Here’s the complete Python code for the implementation:
import numpy as np
# Function to calculate the Multivariate Gaussian Probability
def multivariate_gaussian_probability(x, mean, covariance):
n = len(mean)
diff = x - mean
exponent = -0.5 * np.dot(np.dot(diff.T, np.linalg.inv(covariance)), diff)
coefficient = 1 / ((2 * np.pi) ** (n / 2) * np.linalg.det(covariance) ** 0.5)
return coefficient * np.exp(exponent)
# Example dataset: CPU load and memory usage
data = np.array([[0.5, 1.2], [0.6, 1.4], [0.8, 1.3], [0.7, 1.5], [0.9, 1.7], [0.6, 1.3]])
# Calculate the mean vector and covariance matrix
mean_vector = np.mean(data, axis=0)
covariance_matrix = np.cov(data, rowvar=False)
# Example of a new point
x_new = np.array([1.5, 0.4])
# Calculate the probability using the multivariate Gaussian distribution
probability = multivariate_gaussian_probability(x_new, mean_vector, covariance_matrix)
mean_vector, covariance_matrix, probabilityHere are the results:
I. Parameters of the Multivariate Gaussian Model:
Mean Vector ($\mu$):
- Feature 1 (CPU Load): 0.683
- Feature 2 (Memory Usage): 1.4
Covariance Matrix ($\Sigma$):
[[0.02166667, 0.02 ],
[0.02 , 0.032 ]]II. Probability of New Example: For the new example with a high memory use ($x1 = 1.5$) and low CPU load ($x2 = 0.4$), the calculated probability using the multivariate Gaussian distribution is approximately 8.86e-56.
III. Interpretation:
- The mean vector represents the average CPU load and memory usage from the dataset. This gives the center of the multivariate Gaussian distribution.
- The covariance matrix captures how CPU load and memory usage vary together. A positive covariance between these features suggests that as one increases, the other tends to increase as well.
- The extremely low probability (8.86e-56) indicates that the new example $[1.5, 0.4]$ is highly unlikely under the learned multivariate Gaussian distribution. This combination of high memory use and low CPU load is unusual compared to the typical patterns in the training data, suggesting it could be an anomaly.
Gaussian Model vs. Multivariate Gaussian Model
| Aspect | Gaussian Model | Multivariate Gaussian Model |
| Usage | More commonly used in anomaly detection. | Used less frequently. |
| Feature Creation | Requires manual creation of features to capture unusual combinations in values. | Directly captures correlations between features without needing extra feature engineering. |
| Computational Efficiency | Generally more computationally efficient. | Less efficient computationally. |
| Scalability | Scales well to large feature vectors. | Requires more examples than the number of features (m > n). |
| Training Set Size | Works effectively even with small training sets. | Requires a larger training set relative to the number of features. |
| Advantage | Simple to implement. | Can detect anomalies due to unusual combinations of normal-appearing individual features. |
Reference
These notes are based on the free video lectures offered by Stanford University, led by Professor Andrew Ng. These lectures are part of the renowned Machine Learning course available on Coursera. For more information and to access the full course, visit the Coursera course page.